Implementing genetic algorithm in TypeScript
Now when we know the theory, it 's time to write some code.
Created at:
Last updated:
Now when we know the theory, it 's time to write some code.
Created at:
Last updated:
In this article, we are going to implement a genetic algorithm to solve the traveling salesman problem. If you haven't already, I recommend reading previous articles where I describe what the problem is and what a genetic algorithm is. This background knowledge is necessary to understand what we are coding today.
Let's begin with an analysis of how we can express this problem in terms of code. First, what are the entities we need? So, in terms of a genetic algorithm, we have a population, which represents a pool of individuals in the current iteration. Each individual is represented by its genes.
How can this model work with the problem we are solving? The population has plenty of individuals, each of which has its own genome. This genome is an order of all the cities to visit.
So we can see two entities here: and . We can represent them as TypeScript classes.
We have a simple skeleton here. In the constructor, takes an array of genes. Soon we will add more to it.
In the population right now, we take just a list of individuals on creation.
In addition, we create a class to describe our cities' locations.
From the analysis we've done in previous articles on this topic, we know that the core of a genetic algorithm is the ability to generate new, improved populations by genes crossover and mutation. Crossover works by mixing genes from more than one individual (in our case, two parents), while mutation alters the genome of a single individual.
In my opinion, these two behaviors belong to the , and that's where we are going to put them.
This method will take another individual and return two children created from combined genomes of both parents. The easiest approach is to take a part of the genome from one parent and the other part from the other. In the case of our problem, we cannot just do it like this. Let's look at an example:

We see here two individuals. I also marked a point at which we want to split the first parent's genome. The rest of the genes will come from the other parent. If we simply split the second parent's genome in the same spot and take the second half of it in combination with the first part from the first parent, we made a child. But look at what we have:

At first, it might appear that everything's good, but if we take a closer look, we can see that our child has the city number 1 twice, and it lacks the city number 6. This is not good. We need all the cities and each exactly once.
The better approach is to take the first part of the genome from the first parent as we did, and the rest we fill with cities from the second parent, but only those we don't have yet. Taking our two parents again, the new child will look like this:

Let's look at an implementation:
Here I decided the section point to be in 3/4ths of the genome length. We then create new children - the first one taking the first parent's genome part and filling it with remaining genes from the second parent and the second child the other way around.
Crossover is a mechanism which will create (hopefully) better offspring from existing individuals. This provides us with new populations built from recombined genes from the previous population, but sometimes we might end up in some local optimum, due to the lack of better genes in our population. To overcome this, we can introduce mutation, which will bring some fresh genes to our population, increasing our chance to find an even better solution.
The simplest way to do this is to create a completely random individual once in a while. To control how often we want the mutation to happen we introduce a mutation rate parameter.
That's how we can implement this in the class.
The is a utility function that takes an array and returns a new array with elements randomly placed:
This is all we had to do to have a fully functional class.
Here, things are a bit more complicated. We need to create a new population from the existing one. To generate a new population, we need to create a mating pool - a set of individuals which includes individuals based on their fitness, which is a score of how good the result is for a given individual. Based on this score, the mating pool has more copies of the individuals with higher scores and fewer of those having lower scores.
I think a good approach here is to abstract the fitness logic and introduce a strategy pattern. We want to have a generic fitness strategy interface describing operations we need from the strategy. Our population will take a strategy which fulfills this interface. Now we can implement this interface in different strategy classes and as long as they are compatible with this interface we can pass them to . That way we can have different strategies which are calculating scores differently.
That's how our interface looks like:
And based on that, we can create our own which will give higher scores to individuals with shorter paths.
What we do here is basically calculating the distance between all the cities in the order we get from our individual's genome. Based on that, we have the fitness of an individual.
We also have a which basically adds up all the scores of all individuals we pass as an argument.
Now we want our population to get the fitness strategy as an argument so it can be used to generate new populations. We also want to get the mutation rate, it will be passed to the individuals method.
Now we can move to the heart of our population and add some code responsible for creating a new population for us.
So, we have here a public method which will create a new population. This method first gets a mating pool, which is a new array of individuals. As described before, in this mating pool, individuals appear as many times as good their fitness is. Next, we sort this array by fitness, so in the next step, we cross over pairs of parents next to each other. Before we add an individual to a new population we call on it, passing the mutation rate.
With all this effort, now it is time to use this code to solve the TSP. It would also be nice to see the results. To make it easier, I have created a simple Next.js app which is able to solve the TSP, visualizing its progress over time. The repository can be found here, and this app is also deployed here if you want to play with it.

I know it does not look beautiful, but the purpose of this app was to visualize progress of the algorithm. This simple app allows setting the mutation rate and the amount of cities. We then have two modes of operation - automatic which runs the algorithm in a loop, and manual which runs one iteration. In addition, there is a reset button to start things over with a random set of cities.
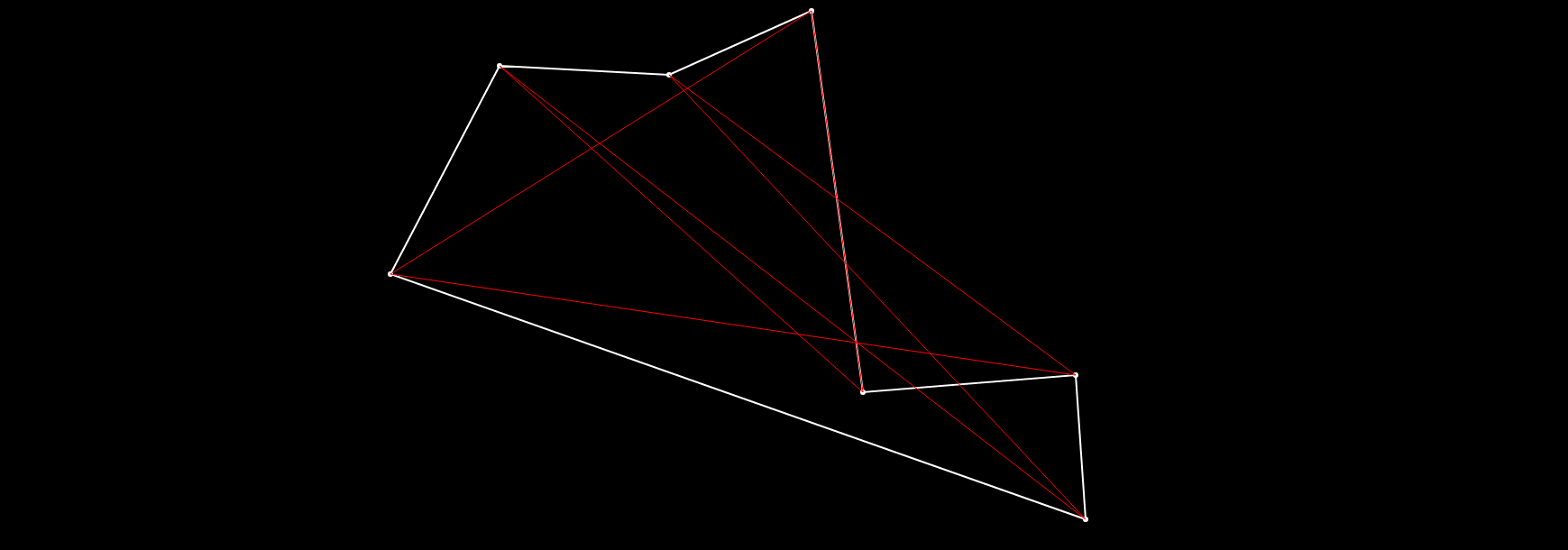
If we run this now, we can see the route drawn in red - this is the worst route in the current population, and a white one - which is the best one.

As you can see, using genetic algorithms isn't that hard. This method of optimization, genetic algorithms, can be applied to virtually any problem. As long as the process we are trying to optimize can be defined by an individual's genome and there is a way to calculate fitness.
I encourage you to play around with parameters, maybe fork my repo and extend its functionality. Try to solve this problem for a higher number of cities, change the population size (it is now hard-coded, but it can be manipulated with an input field).
I hope you enjoyed this short series. Let me know by email if you have any questions or if you want to collaborate on this or any other project.
For now, this is all, and I hope to see you soon in the next one!
Please Sign in to leave a comment.